Clinical surveillance of vector-borne and infectious diseases using molecular diagnostics
Dr. Sufia Sadaf, Program Manager,Public Health Surveillance-Outreach RF-APSI, National Centre for Biological Sciences, NCBS-TIFR, Bangalore

Summary: Public health systems in India are overburdened, with infectious disease incidences on the rise and a lack of rapid and cost-effective public health surveillance system in place. Learning from the SARS-Cov-2 pandemic, several Indian academic and research institutes have set out to conduct focused public health surveillance to prevent disease outbreaks and for better disease preparedness. The Rockefeller Foundation-supported Alliance for Pathogen Surveillance Innovations, APSI-India is a consortium of several academic institutes across four nodal cities in India, Bangalore, Delhi, Hyderabad and Pune which is engaged in public health surveillances, clinical and molecular surveillance of infectious and communicable diseases, environmental surveillance using wastewater and vector surveillances of disease-causing pathogens and those contributing to antimicrobial resistance. Dr. Mansi Malik,Ph.D, Scientist at the Tata Institute for Genetics and Society, India (TIGS), Bangalore, an APSI India partner, has been working towards clinical surveillance of c, and vector borne using molecular methods or molecular tools diseases. In addition, her team explores clinical surveillance of AMR. During the study, her team has created a first-ever combined molecular diagnostic for dengue and another vector borne infection, Chikungunya, which is both early detection of dengue, dengue serotypes and chikungunya and also Denv-Chikv coinfection cost effective and accurate. Through her quest, she has also developed a molecular diagnostic assay to diagnose febrile illnesses to efficiently match it to its causative agents.
Quick glimpse of the article
Dengue, a viral infection in humans, presents with high fever, muscle and joint pain and can be potentially fatal. During the initial 7 days, the viral infection leads to accumulation of immunoglobulin M, IgM and this forms the basis of the serological tests as well as the gold- standard antigen-based serological for its diagnosis, with their own milieu of false positives. Alternatively, cell culture tests to ascertain the virus’s presence can take up to 72 hours for the culture to grow. Because of the false positives and limited duration of the feasibility of serological tests (upto 7 days) and prolonged duration of culture-based diagnosis, molecular test which are based upon nucleic acid amplification, i.e., multiplication of the copies of the viral genetic material, is a more reliable test in recent times. However, these tests cost a fortune and are not affordable by the vast majority of people in India…. Read along for the full story!
 Vector-borne disease, febrile illnesses AMR: Clinical Surveillance at Dr. Mansi Malik’s group, TIGS. Image illustration-Sufia Sadaf
Vector-borne disease, febrile illnesses AMR: Clinical Surveillance at Dr. Mansi Malik’s group, TIGS. Image illustration-Sufia Sadaf Dengue: A vector-borne viral infection
A high fever, headache, rash and muscle and joint pain. As the day progresses, there’s nausea and vomiting. The mind gets dizzy and burdened with panic, of what could this be?
These are the symptoms presented with dengue, and often severe dengue also leads to life- threatening conditions, including bleeding and shock [1, 2].
Dengue is a viral infection, and the causative virus belongs to the genus Flavivirus family, Flaviviridae. The viral infection can be caused by any of its four serotypes (DENV-1, 2, 3, 4) which are antigenically distinct from each other. The disease spreads from infected humans via Aedes mosquitoes to other humans and under most conditions, it causes no symptoms. Often, the moderate symptoms of high fever, headache, muscle and joint pain can be treated with non- inflammatory painkillers. But when it strikes hard, its severe symptoms can be fatal and need hospitalization [1].
Due to the widespread vectors, i.e., the Aedes mosquitoes in tropical and subtropical climates, the dengue disease burden is high. Factors such as climate change, high temperatures/increased humidity, increased financial and health instabilities in countries with humanitarian crises and weakened health systems act as major drivers, and the disease incidence does not seem to stabilize. The final count may still be significantly high because the asymptomatic and mild symptoms often go unreported and are mistaken as a general febrile illness (fever-inducing infections such as those caused by bacterial infections or viral infections, Dengue, Chikungunya, Leptospirosis, Scrubs etc.).
With the endemic nature and its potential to suddenly transition to a life-threatening disease, correct disease diagnosis, pathogen strain identification and disease burden across cities is an absolute requirement for timely and appropriate health intervention and public health policy implementation by the decision makers and government and civic bodies.
With this determination, Dr. Mansi Malik, Scientist at the at the Tata Institute of Genetics and Society (TIGS) Bangalore has been involved in the molecular surveillance of the vector-borne diseases, Dengue and Chikungunya to study the disease burden and hotspots in Bangalore city, the capital of the South Indian state, Karnataka, which contributes to ~50% of the total dengue disease incidence across the country. Her team has extensively studied and developed molecular diagnostic tools to efficiently diagnose these diseases and to distinguish them from other febrile illnesses.
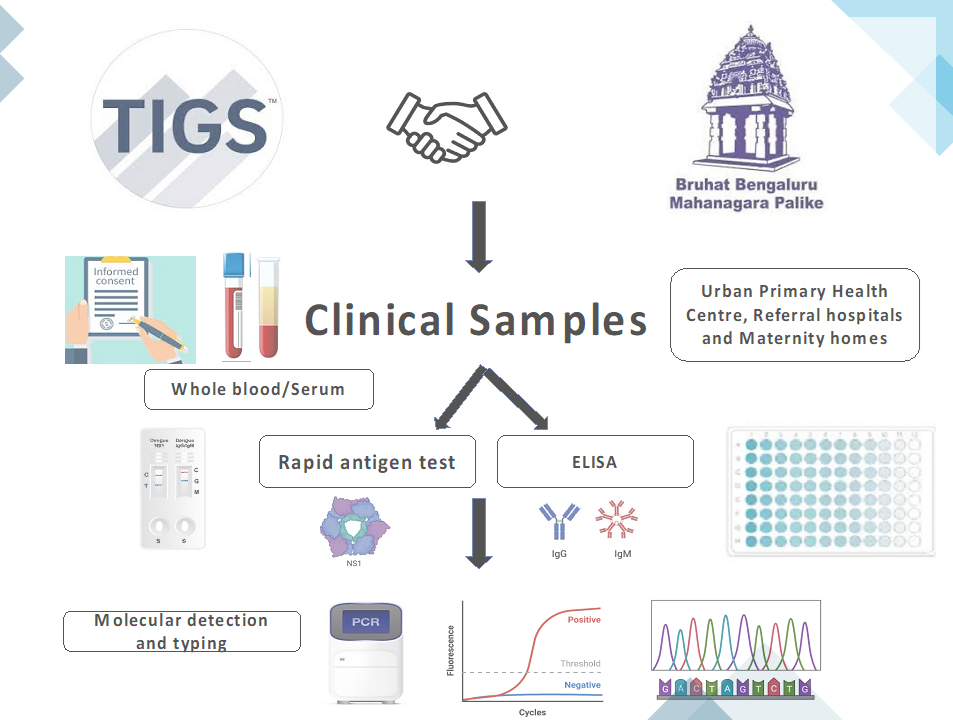 Road map for clinical surveillance across Bangalore city, TIGS. Image illustration-Mansi Malik
Road map for clinical surveillance across Bangalore city, TIGS. Image illustration-Mansi Malik Disease diagnosis: Traditional culture-based and immunology tests
Because the dengue disease agent is a virus, viremia, i.e., the potential to transmit viral infection can last up to 4-5 days or up to 12 days. Disease transmission occurs when the mosquitoes feed on a virus infected human (viremic), the virus incubates in the secondary organs of the insect and after about 12 days, the mosquito turns into a disease transmitting machine for life. It can take up to 2 days for it to transfer the virus to a naive human.
When clinicians/primary health care centers receive a case with the above-mentioned symptoms, most places prescribe the classical diagnostic tests and carry out viral isolation or specialized-cell culture and subsequently serological tests (serum based), by detecting the immunoglobulin, IgM which are produced by the immune system fighting off the viral infection or by probing for the NS1-antigen, a protein produced by dengue virus. The serological tests are conducted via the Enzyme-Linked Immunosorbent Assay (ELISA). Because viral isolation and culture tests can take up to 7 days to yield results and the inability of serological tests to identify/ distinguish between the 4 dengue serotypes and the high magnitude of false positive results, these tests are not rapid and highly efficient in a clinical setting. Alternatively, cell culture and identification of the virus is another mode of diagnosis [3].
 Molecular techniques for Dengue and Chikungunya clinical surveillance. Image illustration-Sufia Sadaf
Molecular techniques for Dengue and Chikungunya clinical surveillance. Image illustration-Sufia Sadaf Due to these limitations, the molecular biology based-rapid amplification of nucleic acid test is more reliable; it involves collecting of viral genetic material, the single stranded ribonucleic acid (RNA) and its amplification through reverse transcription using specific primers, segments of nucleic acid strand which can amplify the target regions with cross-reacting to other dengue serotypes or other pathogen species. The test, referred to as qRT-PCR (quantitative reverse transcriptase polymerase chain reaction) reveals the quantitative assessment of the virus and its various serotypes with an efficiency of up to 99%. The most advanced surveillance comes through genomic sequencing of the virus via NGS (Next generation sequencing) followed by the advanced metagenomics analysis, revealing details of the genome, genes information, mutation prediction, pathogenicity and co-infections.
Despite the reliable efficacy, the nucleic acid tests require a tight deadline! Post infection by day 7, the viral material starts to undergo degradation in the human body, and this creates a need to collect the viral genetic material within the first week of the infection.
On top of the tight timing, most diagnostic centers/ health centers in India charge a hefty amount of money for running the molecular diagnosis (up to ~INR 25000) making them inaccessible to a vast majority of people.
Molecular diagnosis and nucleic acid amplification test
Malik’s team at TIGS, Bangalore conducts clinical surveillance for vector-borne (via mosquito) Dengue and Chikungunya, another viral infection in addition to disease-causing priority pathogens and pathogens contributing to antimicrobial resistance (AMR). The aim of their project is to provide real-time disease burden, hotspots and disease spread statistics in Bangalore city by employing molecular surveillance. Using the resulting data, they study the disease-causing pathogen strains, disease hotspots, disease burden and various co-infections in clinical samples across the city.
Her team receives clinical samples (blood samples) from the city’s primary health centers and other private hospitals. The samples are accompanied by hospital test results ranging from culture-test for bacterial, fungal or plasmodium pathogen positivity and/or with serological ELISA test for the pathogens including viral NS1-antigen or immunoglobulin tests. Next, Malik’s team carries out molecular detection and serotyping (testing for the pathogen strains/ serotypes) which are concordant with the hospital-run assays, rapid (yields results within 1.5 hours) and has up to 95% accuracy compared with 30-40% accuracy of hospital employed routine tests.
By molecular qRT-PCR and genomic sequencing-metagenomic analysis, they can efficiently detect the dengue and chikungunya viral strains and their serotypes even during periods that hospital-run tests are unable to detect positive cases. They further transfer this information to the city public health system, under the BBMP, the local government and civic bodies, for their consideration for timely intervention, policy implementation and informed actions to reduce disease spread.
A novel dual-panel molecular diagnostic kit
As Malik’s team set out to efficiently conduct clinical surveillance of the two vector-borne diseases, Dengue and Chikungunya in Bangalore city, they readily recognized the need to develop strategies to bring down the costs. Through various combinations and pairing of the primers and experimental conditions to run the qRT-PCR tests, they eventually came up with an in-house-dual panel nucleic acid amplification diagnostic kit that could diagnose both Dengue and Chikungunya, along with its various serotypes and co-infections [4]. This offered a one-stop solution to all three problems, by being cost effective, efficient and time saver!
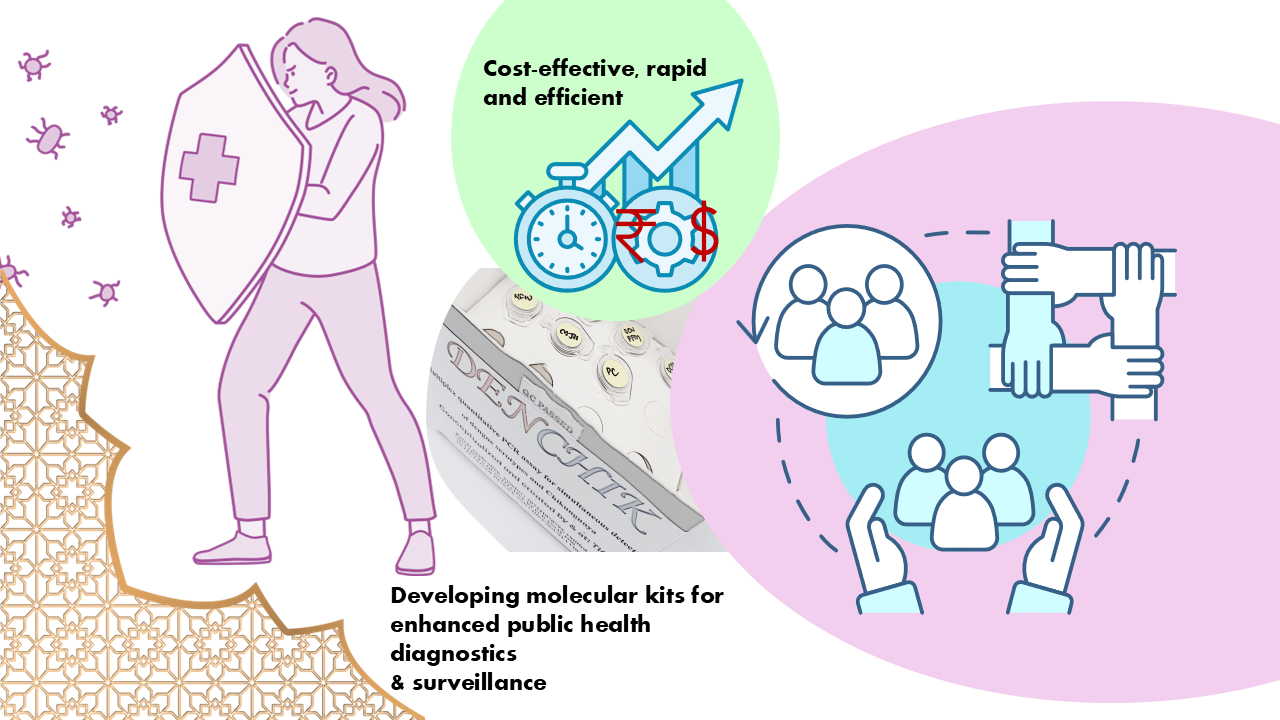 Novel molecular assays and kits for cost-effectiveness and enhanced efficiency. Image illustration-Sufia Sadaf; DENCHIK kit photo- Mansi Malik
Novel molecular assays and kits for cost-effectiveness and enhanced efficiency. Image illustration-Sufia Sadaf; DENCHIK kit photo- Mansi Malik The novel kit, which has been aptly named as DENCHIK has now received a provisional patent under the Govt. of India. Until date, her team has been able to screen over 1500 clinical samples collected for febrile illnesses and have developed molecular diagnostic kits in parallel for pathogens that induce febrile fever, apart from the above described Dengue and Chikungunya (Leptospirosis, Scrub typhus) and viral Hepatitis A,E,C,B. Compared to the skyrocketing prices of the traditional hospital-offered molecular multiplex tests, her team has shown that their in-house tests cost less than INR 600!
So far, they have obtained 4 provisional patents across the molecular diagnostic kits generated to diagnose various disease-causing priority pathogens. Catapulting the public health clinical surveillance towards societal impact and affordability, her team is paving way for the use of molecular techniques to diagnose bacteria, viruses, fungi, as well as plasmodia and to convert these methods to generate novel, low-cost, efficient, in-house kits and expand them further for commercial usage via technology transfer.
Clinical Surveillance of AMR, antimicrobial resistance
Along with the ongoing molecular surveillance, Malik’s team has also been involved in clinical surveillance of pathogens-contributing to antimicrobial resistance (AMR), a leading global and urgent health threat, which contributes to up to 20 million estimated global deaths. Treatment of AMR also incurs huge economic costs and deep financial losses, with an expected additional medical expenditure of $100 trillion by the year 2050 and up to $3.4 trillion gross-domestic losses by the year 2030 [5, 6]!
AMR is induced in bacteria due to excessive usage of antimicrobial drugs, which causes the causative bacteria to develop resistance to the drugs. This leads to a derailed infection control system and a cyclic chain reaction of higher-end drug prescription furthering the induced AMR [5, 6]. The monstrosity of this costly and deadly health condition is rightly termed as a silent tsunami!
Malik’s team has set out to understand gaps in the current diagnostics of AMR and fill the voids with their in-house developed low-cost, high efficacy molecular diagnostic kits. These kits will lead to rapid and accurate acquisition of the results, paving way for a faster and accurate treatment plan, a boon for clinical partners and patients. By combining genomic sequencing and metagenomic analyzed data, her team will also provide in-depth information on the AMR contributing genes in the bacterial genome, antimicrobial resistance genes (ARG), AMR burden, and pathogenicity, pathogen strains and lineage data as well as co-infection possibilities.
 Dr. Malik and her team-at work at TIGS. Novel molecular assays and kit development, along with bioinformatical approaches pathogens, variants and AMR trends for clinical surveillance. Photos-Sufia Sadaf
Dr. Malik and her team-at work at TIGS. Novel molecular assays and kit development, along with bioinformatical approaches pathogens, variants and AMR trends for clinical surveillance. Photos-Sufia Sadaf Her team’s concerted efforts also minimize the cost of labor, all in all being pocket friendly and with the potential to reduce the spread of AMR by removing the aspect of speculative antibiotic prescription. Malik’s team has thus given to society data-backed information, and molecular solutions that will better prepare the public health system against vector borne disease spread and aid in its rapid, cost effective and reliable diagnosis. Malik would like to acknowledge her team at TIGS, Dr. Rakesh Mishra, Director, TIGS, BLISC team, APSI team, her clinical partners and collaborators, BBMP and technology partners for constant support and enthusiastic collaborations!
Congratulations, Malik and team for your achievements and contributions to society! Your relentless efforts have opened avenues for translation of rigorous research to better societal impact and consumption. We wish you many more accomplishments.
Stay tuned for more Science stories from the APSI community!
 Dr. Mansi's group at TIGS. L-R: Prajakta Jathar, Priyadarshini Mohapatra, Mansi Malik, Samruddhi Walaskar, Antony Selvakumar A, Ritika Majji, Priyanka Bhavsar.
Dr. Mansi's group at TIGS. L-R: Prajakta Jathar, Priyadarshini Mohapatra, Mansi Malik, Samruddhi Walaskar, Antony Selvakumar A, Ritika Majji, Priyanka Bhavsar. References
1. Jarman RG et al. Factors influencing dengue virus isolation by C6/36 cell culture and mosquito inoculation of nested PCR-positive clinical samples. Am J Trop Med Hyg, 84(2):218-23 (2011) https://pmc.ncbi.nlm.nih.gov/articles/PMC3029170/
2. WHO Dengue-global situation https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON498
3. Shrivastava S et al. Evaluation of NS1-Detection-Based Cell Culture Method for Isolation of Dengue Viruses from Clinical Samples. SN Compr. Clin. Med. 2, 613–618 (2020). https://doi.org/10.1007/s42399-020-00266-4
4. Uppoor S et al. Development of a cost-effective multiplex quantitative RT-PCR assay for early detection and surveillance of Dengue, Chikungunya, and co-infections from clinical samples in low-resource settings. medRxiv. 24313257 (2024). https://doi.org/10.1101/2024.09.10.24313257
5. WHO_Antimicrobial resistance https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance#:~:text=In%20addition%20to%20death%20and,year%20by%202030%20(2)
6. Financial times: Countries brace for silent tsunami for antibiotic-resistant infections. https://www.ft.com/content/f4c6148a-52e7-4d29-8da0-fe271f3f6a18
—-----------------------------------------------------------------------------
This work and outreach is supported by The Rockefeller Foundation - APSI-India
Article link on LinkedIn: https://www.linkedin.com/pulse/clinical-surveillance-vector-borne-infectious-diseases-using-gjgoc
Visit APSI website: https://data.ccmb.res.in/apsi/
